This section of the website is intended for medical professionals only. To access the client page and online store, please click below.

GHC Genetics to use innovative approach in sequencing oral and intestinal microbiome
Nanopore sequencing belongs to the so-called "third generation" of sequencing, which allows direct reading of single-stranded DNA or RNA without the need for prior amplification (e.g., using PCR) or chemical labeling. The use of nanopores for sequencing was first described in the late 1980s by several American scientists. David Deamer, George Church, and Hagan Bayley independently described the sequencing of a single DNA molecule using electrophoresis and membrane nanopores. This method does not use a fluorescent signal, but measures the voltage changes that occur when DNA passes through an electrically charged pore in the membrane.
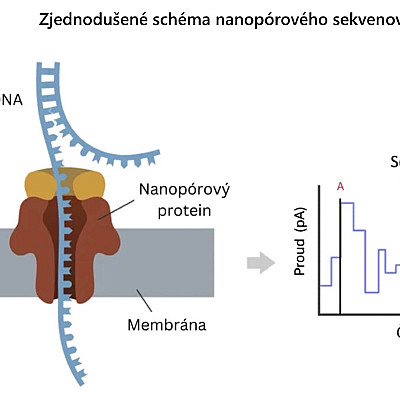
The principle of this method is to embed a biological nanopore into an artificial membrane with a constant electrical resistance, and then apply an electrical voltage to the entire system. The passage and presence of DNA molecules in the nanopore affects the electrical properties of the entire system and causes deviations in the current passing through the nanopore.
The biological membrane with the nanopore is surrounded by an electrolyte solution. The membrane divides the solution into two chambers. Voltage is applied to the membrane and induces an electric field that drives charged particles, in this case ions, into motion. Inside the pore, the DNA molecule occupies a volume that partially restricts the flow of ions, which is observed as a decrease in ion current. Based on various factors, such as geometry, size, and chemical composition, the size of the ion current and the duration of translocation will change (= translocation is a structural rearrangement that occurs when a part of a chromosome is transferred between non-homologous chromosomes). Based on this modulation in the ion current, different molecules can then be detected and potentially identified, with each nucleotide causing a specific voltage change that can be used to determine the sequence.
The main advantage of third-generation methods is their ability to read repetitive sequences, which can range from hundreds of thousands to millions of nucleotides, making them ideal for assembling entire genomes and identifying large genetic changes.
In older methods, the short length of the read segments was a problem because it made it impossible to correctly place the sequences in the genome. This led to difficulties in determining which specific repetitive region a sequence belonged to. Given that the human genome contains more than 50% of such repetitive sequences, third-generation methods are crucial for the analysis of human and other organisms' genomes and contribute to a deeper understanding of biology.
At GHC Genetics, we will soon be using this method to perform sequencing as part of oral and intestinal microbiome testing using the portable MinION MK1D sequencer, which is the size of a cell phone and weighs only 90 grams!